J Pharm Pharmaceut Sci (www.ualberta.ca/~csps) 7(2):205-214, 2004
In vitro characterization and delivery of chitosan-DNA microparticles into mammalian cells.
Tugce Dastan, Kadir Turan1
Department of Pharmaceutical Biotechnology, Faculty of Pharmacy, University of Marmara, Istanbul, TurkeyReceived 18 March 2004, Revised 17 May 2004, Accepted 21 May 2004, Published 30 June 2004
PDF Version
Abstract
PURPOSE: Chitosan has a high potential for transferring DNA molecules into mammalian cells because of its cationic properties. In the present study, we have investigated DNA encapsulation efficiency and loading capacity of chitosan microparticles prepared in different conditions, as well as in vitro DNA release from microparticles, and transfection of different cell lines with chitosan-DNA microparticles, which may be employed in future in vivo studies. METHODS: Plasmid DNAs were amplified in Escherichia coli DH5a and isolated by the alkali SDS-lysis method. Chitosan-DNA microparticles were prepared by the coacervation method by using different concentrations of chitosan and plasmid DNAs. In vitro release experiments were performed in PBS at 37°C and DNA release was monitored spectrophotometrically. Transfection efficiency of chitosan-DNA microparticles into mammalian cells was determined by measuring the b-galactosidase activity in cell lysates. RESULTS: DNA encapsulation efficiency and loading capacity of microparticles was altered depending on the chitosan and DNA concentrations. Approximately 75-85% of DNA was encapsulated into the chitosan-DNA microparticles. The average size of microparticles was found to be ~2 mm. In vitro studies revealed that the release of DNA from chitosan microparticles could be controlled by changing the formulation conditions. Although the transfection efficiency of chitosan-DNA microparticles was typically lower than that of DNA complexed with lipid-based reagents, in vitro transfection results indicated that HEK293 cells take up chitosan-DNA microparticles more efficiently compared to HeLa and mouse fibroblastic 3T3 cell lines. Conclusion: Chitosan microparticles provide a sustained release of plasmid DNA for a long period and they have a potential for DNA transfer into the mammalian cells. However, transfection efficiency of chitosan-DNA microparticles is low and dependent on the cell type.
Introduction
Chitosan is a deacetylated derivative of chitin, which is one of the naturally most abundant mucopolysaccharides and supporting materials of crustaceans and insects. It is a non-toxic and easily biodegradable polymer (1-3). Polymeric controlled delivery systems based on chitosan are now being used for a wide range of drugs (4-6). Chitosan as a non-viral vehicle for transferring DNA molecules into the cells has recently attracted much attention because of its unique properties (7, 8).
The efficient delivery of functional therapeutic genes into target cells in vitro and in vivo is an important problem in gene therapy approaches for the treatment of hereditary diseases and cancer (9, 10). Various methods have been developed to accomplish gene transfer into eukaryotic cells. These methods involve the use of genetically modified viruses to deliver the genetic material (11, 12), non-viral gene transfers including the direct physical introduction of the genetic material into cells (13-15) and formation of DNA complexes with inorganic salts, polycations, or lipids to transfer the DNA into the cells (16-18). One of the major gene transfer vehicles, genetically modified viruses such as retrovirus, adenovirus, etc., have been utilized for approximately 80% of approved phase I clinical trials because of ease of transfer of their genome into the cells (19). However, they have some disadvantages in their usage in vivo such as immunogenicity, insertional mutations, and potential pathogenicity (20). In contrast, a non-viral delivery system has several advantages including low cost, non-infectivity, absence of immunogenicity, and the possibility of repeated clinical administration (21). Therefore, the research and development of non-viral vectors is a very important alternative approach in gene therapy.
In recent years, the potential of chitosan as a non-viral gene carrier has been extensively considered by several research groups. Chitosan effectively binds to negative surfaces because of its positive charge (22). At acidic pH, the amine groups of chitosan become positively charged, bind to DNA and condense it as nano/microparticles (23-26). The size and shape of particles can be changed depending on the conditions of the formulation process (26, 27). Chitosan microparticles containing reporter genes are being used for the transfection of mammalian cells both in vitro and in vivo conditions (28-30). However, the transfection efficiency of microparticles is quite low. On the other hand, extensive efforts are being expended for improving new non-viral vehicles by using chitosan polymers (31). Different approaches including the modification of chitosan by adding some groups (32-34), conjugation of some specific ligands to chitosan nano/microparticles (27), and formulation of DNA by using the combination of chitosan and other polymers like poly-L-lysine (35-37) are being applied for enhancing the transfection efficiency and targeting DNA as chitosan nano/microparticles. Several derivatives of chitosan have been prepared based on the reaction with the free amine groups (38, 39). Kim et al. (32) have recently prepared a galactosylated chitosan to formulate plasmid DNA as nanoparticles in order to improve the transfection efficiency and targeting chitosan-DNA nanoparticles and, they showed that HepG2 (human hepatoblastoma cell line) having asialoglycoprotein receptors binding galactose-containing ligands was more efficiently transfected than HeLa without asialoglycoprotein receptors. On the other hand, it was reported that lactosylated-chitosan/DNA complexes efficiently transfected HeLa cells, but not HepG2 (40). Viruses have excellent strategies for transferring their genome into the host cells. Viral infection process is a very useful model for improved non-viral vectors. Mao et al. (27) have conjugated KNOB (C-terminal domain of the fiber protein of adenoviruses) to chitosan-DNA nanoparticles and reported a 130-fold increase in transfection efficiency in HeLa cells. In contrast, a low level of transfection efficiency was observed with chitosan-DNA nanoparticles conjugated with transferrin (a ligand having a role in the transfer of small molecular weight drugs and bioactive macromolecules into mammalian cells, 41) in HEK293 and HeLa cells (27). Although there are several reports supporting the use of chitosan as a non-viral vector, studies regarding improvements in transfection efficiency and controlled expression of related gene remain insufficient. In this respect, we have formulated plasmid DNAs as chitosan-DNA microparticles and investigated in vitro release profiles of DNA from these microparticles. We have also assayed the transfection capability of chitosan-DNA microparticles into different mammalian cells.
Methods
Materials
Chitosan, 400 kD MW and 87% deacetylated, was purchased from Fluka Co. Ltd., Germany. The cell culture media and reagents were purchased from Gibco, USA. All other chemicals used were molecular biology and pharmaceutical grade.
Plasmids
Three different sizes of plasmids were used. pSVβ-Gal (Promega, USA) containing bacterial b-galactosidase gene under the control of SV40 promotor is 6820 bp. pEGFP-N1 (Clontech, USA) containing jelly fish green fluorescent protein gene under the control of CMV immediate early promoter and pBluescript Π SK+ (MBI Fermentas, Litvanya) are 4733 bp and 2961 bp, respectively. Escherichia coli DH5a was used as host cell for amplification of plasmids. Plasmid DNAs were isolated with alkali-lysis method and purified with CsCl-EtBr banding.
Encapsulation of plasmid DNA in chitosan microparticles
Chitosan microparticles were prepared by the coacervation method (42). Chitosan was dissolved in 2% acetic acid solution at 0.25%, 0.50% or 0.75% (w/v) final concentrations. DNA samples were dissolved in 20% sodium sulfate solution at 5-40 ìg/ml final concentrations. For the formation of chitosan-DNA microparticles, an equal volume of sodium sulfate solution containing DNA was added dropwise to the chitosan solutions and mixed on a magnetic stirrer for 1 hour. Chitosan-DNA microparticles were recovered by centrifugation and after washing with distilled water, freeze-dried or dialyzed against distilled water. The loading degree was determined by quantifying the non-bound DNA in supernatant with a spectrophotometer. Both loading capacity (LC) and loading efficiency (LE) were calculated by using the following equations: LC (%) = [(total amount of DNA) - (non-bound DNA) / weight of chitosan-DNA microparticles] x 100; LE (%) = [(total amount of DNA) - (non-bound DNA) / total amount of DNA] x 100..
Particle size analysis
The size of chitosan-DNA microparticles and its polydispersity were analyzed from light microscopy images of microparticles by using Kodak 1D 3.5 software. The microparticles were also examined with transmission electron microscopy (TEM) (Zeiss EM9A). For TEM examination, the sample was placed on a formvar coated copper grid and negatively stained by an aqueous solution of ammonium molibdate (1% w/v).
In vitro release of plasmid DNA
Chitosan-DNA microparticles (20 mg) were suspended in 4 ml of phosphate buffered saline (pH 7.4). The suspension was shaken in a water bath at 37ºC. At defined time intervals, the supernatant was collected by centrifugation and microparticles were re-suspended in fresh buffer. Plasmid DNA released into the supernatant was quantified by measuring with a spectrophotometer.
Analysis of plasmid DNA released from chitosan microparticles
For the determination of integrity of plasmid DNA encapsulated in chitosan microparticles, DNA samples released from microparticles were analyzed with agarose gel electrophoresis. Before loading onto the gel, DNA samples were concentrated by precipitating with ethanol and passed through Sephadex G-25 gel filtration and DEAE-Sepharose CL-6B cationic-exchange columns. Column fractions were collected in 500 ml volume and 20 ml from each fraction were loaded onto the 1% agarose gel.
Cell lines
Human embryonic kidney (HEK293), Swiss3T3, and HeLa cell lines were used for transient transfection experiments. HEK293 cells were maintained at 37ºC under 5% CO2 in Dulbecco's modified Eagle's medium-high glucose (D'MEM-HG) (Gibco) supplemented with 10% (v/v) fetal calf serum (Gibco), penicillin (100 U/ml) and streptomycin (100 mg/ml).
The other cell lines were maintained in the same conditions as HEK293 cells except Dulbecco's modified Eagle's medium containing low glucose.
Transfection of cells and β-galactosidase assay
Cells were seeded in 24-well plates (5 x 104 cells/well) and grown at standard culture conditions for 24 hours. Culture media were changed with fresh complete media containing the defined concentration of chitosan-DNA microparticles.
After 48 hours incubation, cells were harvested for b-galactosidase assay. Briefly, culture media were discarded and the cells were washed with PBS.
The cells were detached with trypsin, suspended in PBS, and collected by centrifugation. The cells were lysed in 200 ml of lyses buffer containing 100 mM KH2 PO4 / K2 HPO4 (pH 7.5), 0.2% Triton X-100, 1 mM DTT by freezing and thawing. The b-galactosidase assay was performed in a microtiter dish. Twenty-five ìl of cell lysate was added to 135 ml of buffer A containing 100 mM KH2 PO4 / K2 HPO4 (pH 7.5), 10 mM KCl, 1 mM MgSO4, and 50 mM 2-mercaptoethanol, and incubated for 5 minutes at 37ºC. Then, 50 ml ONPG (O-nitrophenyl-b-D-galactopyranoside) substrate solution (4 mg/ml ONPG in 100 mM phosphate buffer, pH 7.5) was added to the reaction mixture and incubated for 1-16 h at 37ºC. After the incubation period, the reaction was terminated by addition of 90 ml stop solution (1 M Na2 CO3 ) and the absorbance of samples was measured with a microtiter dish reader set at 420 nm. Protein concentration of cell lysate was determined with Bradford method.
The b-galactosidase activity was calculated by using the following equations and units of enzyme were expressed as nmoles of b-galactose formed per minute (modified from ref. 43). b-galactosidase activity (U/mg of total protein in lysate) = [OD420 /0.0045 x assay volume (ml)].min 1 .mg-1.
Results and Discussion
The chitosan-DNA microparticles formed by coacervation method were examined in terms of their size, DNA loading efficiency and DNA loading capacity (Table 1).
Table 1: The properties of chitosan-DNA microparticles made with different concentration/size of plasmid DNA and different concentration of chitosan. Data are expressed as mean ± SD (n=5 for LE and LC, n=500 for microparticles sizes).
The loading efficiency of chitosan-DNA microparticles increased depending on the concentration of chitosan solution. Approximately 75-85% of total DNA was encapsulated into the microparticles. In contrast, the loading capacity of microparticles decreased depending on the concentration of chitosan solution.
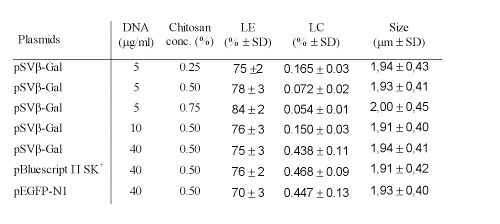
The size distribution of microparticles was found to be between 1.3-3.3 mm (Fig 1).
Figure 1: Particle size distributions of chitosan-DNA microparticles.
The average particle size of different chitosan-DNA formulations was found to be ~2 mm. There were no significant differences in size and polydispersity among the chitosan microparticles prepared by using different concentration of chitosan solutions.
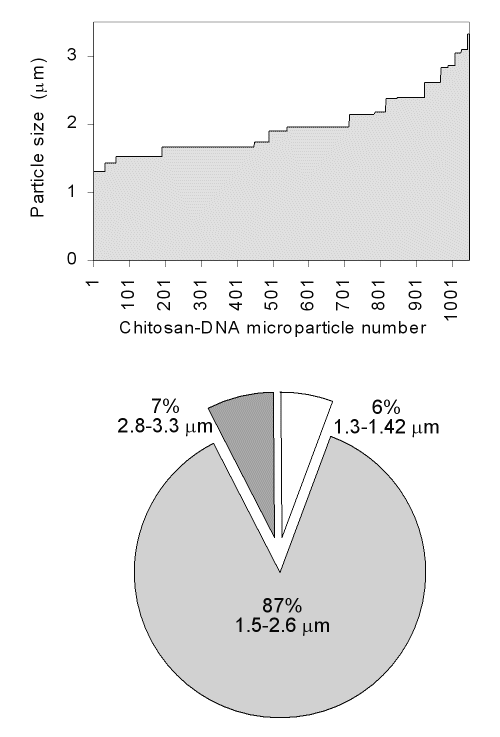
Individual microparticles with a sphere-like shape or clustered groups were observed by light microscopy and TEM (Fig 2).
Figure 2: Light microscopy (A) and TEM (B) images of chitosan-DNA microparticles.
The chitosan-DNA microparticles described in this study were smaller than 5 mm, being a suitable size for mammalian cells uptake.
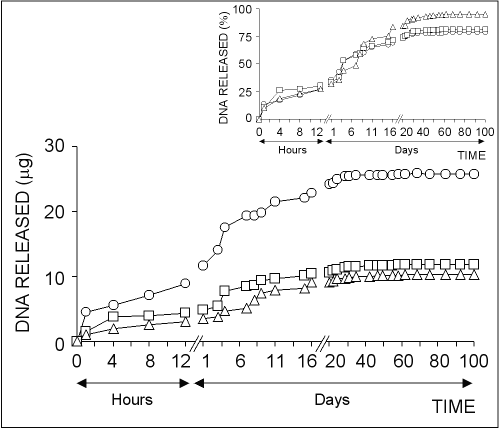
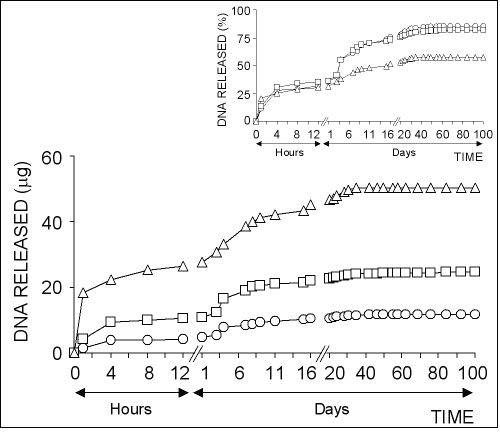
The in vitro plasmid DNA release from chitosan microparticles formed by using different concentration of chitosan solutions is illustrated in Fig 3.
Figure 3: The effect of chitosan concentration on the release of pSVb-Gal plasmid DNA. Chitosan-DNA microparticles were prepared by mixing equal volume of 0.25% (ο), 0.50% () or 0.75% (∆) of chitosan solutions and 20% of sodium sulfate solution containing 5 mg/ml pSVb-Gal plasmid DNA. Small figure shows the amount of DNA released in percentage values that are calculated from the loading capacity of chitosan-DNA microparticles.
About 10-15% of total loaded DNA into the chitosan microparticles was initially released within the first one hour followed by very slow release over 40-50 days.
The highest amount of DNA was released from chitosan-DNA microparticles formed with 0.25% chitosan solution.
In contrast, the highest DNA release percentage was observed in microparticles formed with 0.75% chitosan solution (Fig 3, small figure).
About 95% of the loaded DNA was released from chitosan microparticles prepared with 0.75% of solution at the end of 100 days experiment period, while the ratio was realized as ~80% for chitosan microparticles prepared with 0.25% and 0.50% chitosan solutions.
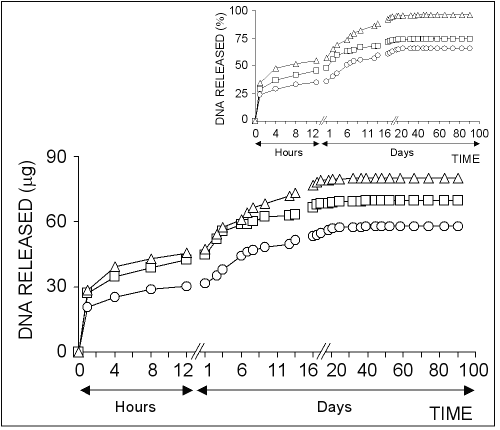
The effect of plasmid DNA concentration on release rate from chitosan microparticles was examined as shown in Fig 4.
Figure 4: The effect of DNA concentration on the release of pSVb-Gal plasmid DNA. Chitosan-DNA microparticles were prepared by mixing equal volume of 0.50% of chitosan solution and 20% of sodium sulfate solutions containing 5 mg/ml (ο), 10 mg/ml () or 40 mg/ml (Ä) of pSVb-Gal plasmid DNA. Small figure shows the amount of DNA released in percentage values.
It was shown that the DNA release rate from microparticles was increased depending on the concentration of plasmid DNA used for the formation of chitosan microparticles.
In contrast, the percentage release of the loaded DNA decreased depending on the DNA concentration.
About 60% of the loaded DNA was released from chitosan microparticles prepared by using 40 ìg/ml DNA solution (Fig 4, small figure).
These results suggest that the ratio of chitosan and DNA used for the formation of chitosan-DNA microparticles is highly effective on DNA release profiles.
Another factor affecting the DNA release from microparticles is the size of DNA molecules encapsulated in the chitosan microparticles.
Fig 5 shows the release profiles of different size of plasmid DNAs from chitosan microparticles prepared in the same conditions.
Figure 5: The profiles of DNA released from chitosan microparticles loaded with different size of plasmids. Chitosan-DNA microparticles were prepared by mixing equal volume of 0.50% of chitosan solution and 20% of sodium sulfate solutions containing 40 mg/ml of pSVb-Gal (ο), pEGFP-N1 () or pBluescript Π SK + (∆). Small figure shows the amount of released DNA in percentage values.
The results showed that the higher molecular weight plasmids were released slower than that of smaller. Electrophoretic analysis of the released DNA showed that DNA molecules encapsulated in chitosan microparticles were kept intact against degradation (Fig 6).
Figure 6: Electrophoretic analysis of pSVb-Gal DNA released from chitosan-DNA microparticles. Panel A: Elution profiles of released plasmid DNA after concentrating with ethanol precipitation. DNA samples were eluted with deionized distilled water from Sephadex G-25 column. DNA samples were loaded on DEAE-Sepharose ion-exchange column with 0.1 M NaCl and eluted with 1 M NaCl solutions; Panel B: Electrophoretic profiles of plasmid DNA eluted from Sephadex G-25 gel filtration column. Lane M: molecular weight marker (Lambda phage DNA/HindIII and EcoRI digested), Lane K: intact pSVb-Gal, Lanes 2-8: column fractions. Panel C: Electrophoretic profiles of the plasmid DNA eluted from DEAE-Sepharose ion exchange column. Lanes 3-9: column fractions.
Before loading onto the agarose gel, released DNA samples were concentrated and passed through column chromatograph. The elution profiles of DNA samples are given in Fig 6, panel A. Sephadex G-25 gel filtration column was found to be inefficient for the separation of DNA from chitosan contaminants. It is known that cationic chitosan polymers form complex with DNA molecules and affect the electrophoretic mobility of DNA by changing the net electrical charge (26). A similar result was observed in our experiments. Plasmid DNA in fractions eluted from Sephadex G-25 column immigrated on agarose gel (Fig 6, panel B). In contrast, released plasmid DNA was sufficiently purified with DEAE-Sepharose cationic-exchange column for electrophoretic analysis (Fig 6, panel C). Electrophoretic profiles of DNA purified with DEAE-Sepharose indicated that the encapsulation and release processes were not detrimental on the integrity of plasmid DNA.
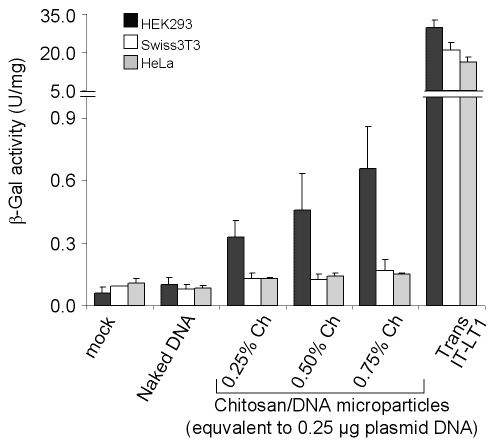
Transfection of mammalian cells with chitosan-DNA microparticles was established by using â-galactosidase expressing plasmid, pSVb-Gal.
Although Swiss3T3 and HeLa cell lines were found resistant to transfection with chitosan microparticles, HEK293 cells were transfected more efficiently (Fig 7).
Figure 7: Transfection of mammalian cells with pSVb-Gal as naked DNA, formulated in chitosan microparticles or complexed with a commercial transfection reagent. Chitosan-DNA microparticles were dialyzed in deionized distilled water after formation by coacervation method. The cells were grown in 24-well plate and transfected with 0.25 mg of pSVb-Gal DNA or 0.25 mg of pSVb-Gal equivalent chitosan-DNA microparticles. Naked plasmid DNA was added into the cultures without any processing. Transfection with Trans IT-LT1 was carried out according to manufacturer's instructions. Error bars represent standard deviation (n = 3).
However, transfection efficiency of HEK293 cells with chitosan-DNA was very low compared to the lipid-based commercial transfection reagent Trans IT-LT1 (Mirus). The presence or absence of serum in the transfection medium did not affect the transfection efficiency (data not shown). This result indicated that different cell lines show different transfection efficiencies with chitosan-DNA microparticles.
Chitosan is a non-toxic and biodegradable polymer and easily formulated as nano/microparticles, beads or membranes (1). In this study, we formulated plasmid DNAs as chitosan-DNA microparticles and revealed their in vitro release properties. After 10-15% initial release in the first one hour, a sustained release of plasmid DNA was observed. In principle, plasmid DNA exists in three isoforms including supercoiled, open circular and linear (44). It has a higher molecular weight compared to most of the bioactive molecules and it forms stable complexes with polycationic chitosan. In the studies carried out with low molecular weight drugs like ampicillin (45), doxorubicin (46) and diclofenac sodium (47) formulated as chitosan nano/microparticles, it was observed that the release of drug was completed in hours. In contrast, DNA release from chitosan microparticles continues several months. In this study we showed that released DNA from chitosan microparticles do not migrate in agarose gel without purification with DEAE-Sepharose ion exchange column (data not shown and Fig 6B). These results suggest that the DNA molecules are released from microparticles because of surface erosion rather than diffusion.
One of the important factors affecting the expression level of a gene located on transfected DNA is its copy number. Therefore, chitosan-DNA microparticles have the potential of intracellular sustained release of DNA that might be beneficial for a more prolonged and controlled expression of gene product. On the other hand, DNA molecules can be formulated with chitosan polymers under mild conditions. Chitosan polymers protect DNA molecules from enzymatic digestion both in vivo and in vitro conditions (36, 37, 48). The results from the electrophoretic studies showed that encapsulation and release processes are not destructive to plasmid DNA. This may improve the bioavailability of plasmid DNA for in vivo applications. Our in vitro transfection results showed that the cell type is an important limitation for DNA delivery with chitosan polymers. HEK293 cells were more efficiently transfected than HeLa and fibroblastic 3T3 cells. Chitosan-DNA microparticles prepared with 0.75% chitosan solution more effectively transfected HEK293 cells. This result may depend on the relative excess amount of chitosan microparticles containing 0.25 mg equivalent plasmid DNA.
Chitosan-DNA nanoparticles have been reported to transfect various cell types, mainly Hela, HEK293 and COS-1 cells (23, 49). However, it was shown that the cell type is important for DNA transfection with chitosan polymers (28, 40). Leong et al (24) reported the HEK293 cells were more efficiently transfected than Hela with chitosan-DNA nanoparticles. Whereas, it was demonstrated that there is a 130-fold increase in transfection efficiency of HeLa cells by conjugation of DNA-chitosan nanoparticles with KNOB (27). Corsi et al. (28) also reported a higher transfection efficiency of HEK293 than M69 mesenchymal cell line with chitosan nanoparticles. In present study, we showed that HEK293 cells were transfected with chitosan-DNA microparticles, while HeLa and Swiss 3T3 cells were resistant to transfection, being in agreement with previous reports. All these results suggest that the composition of cell membrane might be an important factor in adsorption and internalization of chitosan-DNA particles. However, the adsorption and internalization of chitosan-DNA particles are not only limiting factors in gene expression (10). Ishii at al. (26) showed that chitosan-DNA complexes were internalized into SOJ cell by endocytosis and accumulated inside the nucleus as complexes. It was also reported that chitosan-DNA complexes were internalized by HeLa cells and located inside endosomes (40). Consequently, the low level of transfection efficiency of mammalian cells with chitosan-DNA particles may also be a result of formation of strong complexes between chitosan polymers and DNA, which interfere with the transport of genes into the nucleus or their transcription.
In conclusion, we formulated DNA molecules as chitosan-DNA microparticles by using coacervation method and revealed that DNA molecules release from microparticles for a long period in vitro condition, which may provide an intracellular sustained release of DNA in vivo. Different cell types showed different transfection efficiency with chitosan-DNA microparticles. Consequently, the cell type used for gene transfer with chitosan polymers and formulation conditions should be considered in vivo applications.
References
Janes, K. A., Calvo, P., and Alonso, M. J., Polysaccharide colloidal particles as delivery systems for macromolecules. Adv Drug Deliver Rev, 47: 83-97, 2001.
Kas, H. S., Chitosan properties, preparations and application to microparticulate systems. J Microencapsul, 14: 689-711, 1997.
Kumar, M. N. V. R., A review of chitin and chitosan applications, React Funct Polym, 46: 1-27, 2000.
Lee, J. Y., Nam, S. H., Im, S. Y., Park, Y. J., Lee, Y. M., Seol, Y. J., Chung, C. P., and Lee, S. J., Enhanced bone formation by controlled growth factor delivery from chitosan-based biomaterials. J Cont Rel, 78: 187-197, 2002.
Janes, K. J., Fresneau, M. P., Marazuela, A., Fabra, A., and Alonso, M. J., Chitosan nanoparticles as delivery systems for doxorubicin. J Cont Rel, 73: 255-267, 2001.
Singla, A. K., and Chawla, M., Chitosan: some pharmaceutical and biological aspects - an update. J Pharm Pharmacol, 53: 1047-1067, 2001.
Borchard, G., Chitosans for gene delivery. Adv Drug Deliver Rev, 52: 145-150, 2001.
Liu, W. G., and Yau, K. D., Chitosan and its derivatives - a promising non viral vector for gene transfection, J Cont Rel, 83: 1-11, 2002.
Ruponena, M., Honkakoskia, P., Ronkko, S., Pelkonenb, J., Tammic, M., and Urtti, A., Extracellular and intracellular barriers in non-viral gene delivery. J Cont Rel, 93: 213-217, 2003.
Ferrari, S., Geddes, D., M., and Alton, E. W. F. W., Barriers to and new approaches for gene therapy and gene delivery in cystic fibrosis, Adv Drug Deliver Rev, 54: 1373-1393, 2002.
Lundstrom, K., Latest development in viral vectors for gene therapy. Trends Biotechnol, 21: 117-122, 2003.
Pana, Y., Zhaia, P., Dashtia, A. M., Wua, S., Lina, X., and Wub, M., A combined gene delivery by co-transduction of adenoviral and retroviral vectors for cancer gene therapy. Cancer Lett, 184: 179-188, 2002.
Suzuki,T., Shin, B. C., Fujikura, K., Matsuzaki, T., and Takata, K., Direct gene transfer into rat liver cells by in vivo electroporation. FEBS Lett, 425: 436-440, 1998.
Brown, M. D., Schatzlein, A. G., and Uchegbu, I. F., Gene delivery with synthetic (non viral) carriers. Int J Pharm, 229: 1-21, 2001.
Murphy, R. C., and Messer, A., Gene Transfer Methods for CNS Organotypic cultures: A comparison of three nonviral methods. Mol Ther, 3: 113-121, 2001.
Kumar, M. N. V. R., Bakowsky, U., and Lehr, C. M., Preparation and characterization of cationic PLGA nanospheres as DNA carriers. Biomaterials, 25: 1771-1777, 2004.
Almofti, M. R., Harashima, H., Shinohara, Y., Almofti, A., Baba, Y., and Kiwada, H., Cationic liposome-mediated gene delivery: Biophysical study and mechanism of internalization. Arch Biochem Biophys, 410: 246-253, 2003.
Reschel, T., Konak, C., Oupicky, D. Leonard W. Seymour, L. W., and Ulbrich, K., Physical properties and in vitro transfection efficiency of gene delivery vectors based on complexes of DNA with synthetic polycations. J Cont Rel, 81: 201-217, 2002.
Mountain, M., Gene therapy: the first decade. TIBTECH, 18: 119-128, 2000.
Stone, D., David, A., Bolognani, F., Lowenstein, P. R., and Castro, M. G., Viral vectors for gene delivery and gene therapy within the endocrine system. J Endocrinol, 164: 103-18, 2000.
Pouton, C. W., and Seymour, L. W., Key issues in non-viral gene delivery. Adv Drug Deliver Rev, 34: 3-19, 1998.
Hayatsu, H., Kubo, T., Tanaka, Y., and Negishi, K., Polynucleotide-chitosan complex, an insoluble but reactive form of polynucleotide. Chem Pharm Bull, 45: 1363-1368, 1997.
Lee, K. Y, Kwon, I. C., Kim, Y. H., Jo, W. H., and Jeong, S. Y., Preparation of chitosan self-aggregates as a gene delivery system. J Cont Rel, 51: 213-220, 1998.
Leong, K. W., Mao, H. Q., Truong-Le, V.L., Roy, K., Walsh, S.M., and August, J. T., DNA-polycation nanospheres as non-viral gene delivery vehicles. J Cont Rel, 53: 183-193, 1998.
Maclaughlin, F. C., Mumper, R. J., Wang, J., Tagliaferri, J. M., Gill, I., Hinchcliffe, M., and Rolland, A. P., Chitosan and depolymelized chitosan oligomers as condensing carriers for in vivo plasmid delivery. J Cont Rel, 56: 259-272, 1998.
Ishii, T., Okahata, Y., and Sato, T., Mechanism of cell transfection with plasmid/chitosan complexes. Biochim Biophys Acta, 1514: 51-64, 2001.
Mao, H. Q., Roy, K., Troung-Le, V. L., Janes K. A., Lin, K. Y., Wang, Y., August, J. T., and Leong, K. W., Chitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiency. J Cont Rel, 70: 399-421, 2001.
Corsi, K., Chellat, F., Yahia, H., and Fernandes, J. C. Mesenchymal stem cells, MG63 and HEK293 transfection using chitosan-DNA nanoparticles. Biomaterials, 24: 1255-1264, 2003.
Sato, T., Ishii, T., and Okahata, Y., In vitro gene delivery mediated by chitosan. Effect of pH, serum, and molecular mass of chitosan on the transfection efficiency. Biomaterials, 22: 2075-2080, 2001.
Iqbal, M., Lin, W., Jabbal-Gill, I., Davis, S. S., Steward, M. W., and Illum, L., Nasal delivery of chitosan–DNA plasmid expressing epitopes of respiratory syncytial virus (RSV) induces protective CTL responses in BALB/c mice. Vaccine, 21: 1478-1485, 2003.
Mansouri, S., Lavigne, P., Corsi, K., Benderdour, M., Beaumont, E., and Fernandes, J. C., Chitosan-DNA nanoparticles as non-viral vectors in gene therapy: strategies to improve transfection efficacy. Eur J Pharm Biopharm, 57: 1-8, 2004.
Kim, T. H., Park, I. K., Nah, J. W., Choi, Y. J., and Cho, C. S., Galactosylated chitosan/DNA nanoparticles prepared using water-soluble chitosan as a gene carrier. Biomaterials, 25: 3783-3792, 2004.
Park, I. K., Kim, T. H., Park, Y. H., Shin, B. A., Choi E. S., Chowdhury, E. H., Akaike, T., and Cho, C. S., Galactosylated chitosan-graft-poly(ethylene glycol) as hepatocyte-targeting DNA carrier. J Cont Rel, 76: 349-362, 2001.
Thanou, M., Florea, B. I., Geldof, M., Junginger, H. E., and Borchard, G., Quaternized chitosan oligomers as novel gene delivery vectors in epithelial cell lines. Biomaterials, 23: 153-159, 2002.
Aral, C., and Akbuga, J., Preparation and in vitro transfection efficiency of chitosan microspheres containing plasmid DNA: poly(L-lysine) complexes, J Pharm Pharm Sci, 6: 321-326, 2003.
Quong, D., Yeo, J. N., and Neufeld, R. J., Stability of chitosan and poly-L-lysine membranes coating DNA-alginate beads when exposed to hydrolytic enzymes. J Microencapsul, 16: 73-82, 1999.
Quong, D., and Neufeld, R. J., DNA protection from extracapsular nucleases, within chitosan- or poly-L-lysine-coated alginate beads. Biotechnol Bioeng, 60: 124-34, 1998.
Lillo, L. E., and Matsuhiro, B., Chemical modification of carboxylated chitosan. Carbohydr polym, 34: 397-401, 1997.
Xu, Y., Du, Y., Huang, R., and Gao, L., Preparation and modification of N-(2-hydroxyl) propyl-3-trimethyl ammonium chitosan chloride nanoparticle as a protein carrier. Biomaterials, 24: 5015-5022. 2003.
Erbacher, P., Zou, S., Bettinger, T., Steffan, A. M., and Remy, J. S., Chitosan-based vector/DNA complexes for gene delivery: biophysical characteristics and transfection ability. Pharm Res, 15: 1332-1339, 1998.
Cheng, P. W., Receptor ligand-facilitated gene transfer: enhancement of liposome-mediated gene transfer and expression by transferrin. Hum Gene Ther, 7: 275-282, 1996.
Berthold, A., Cremer, K., and Kreuter J., Preparation and characterization of chitosan microspheres as drug carrier for prednisolone sodium phosphate as model for anti-inflammatory drugs. J Cont Rel, 39: 17-25, 1996.
Nielsen, D. A., Chou, J., MacKrell, A. J., Casadaban, M. J., and Steiner, D. F., Expression of a preproinsulin-beta-galactosidase gene fusion in mammalian cells. Proc Natl Acad Sci U S A, 80: 5198-5202, 1983.
Summers, D., K., The biology of plasmids. Blackwell Science, USA, 1996.
Giunchedi, P., Genta, I., Conti, B., Muzzarelli, R. A. A., and Conte, U., Preparation and characterization of ampicillin loaded methylpyrrolidinone chitosan and chitosan microspheres. Biomaterials, 19: 157-161, 1998.
Kevin A. J., M. P., Fresneau, A. Marazuela, A. Fabra, and M. J., Alonso, Chitosan nanoparticles as delivery systems for doxorubicin. J Cont Rel, 73: 255-267, 2001.
Gupta, K. C., and Kumar, M. N. V. R., Drug release behavior of beads and microgranules of chitosan. Biomaterials, 21: 1115-1119, 2000.
Richardson, S. C. W., Kolbe, H. V. J., and Duncan, R., Potential of low molecular mass chitosan as a DNA delivery system: biocompatibility, body distribution and ability to complex and protect DNA. Int J Pharm, 178: 231-243, 1999.
Kiang, T., Wen, J., Lim, H. W., and Leong, K. W., The effect of the degree of chitosan deacetylation on the efficiency of gene transfection. Biomaterials, 25: 5293-5301, 2004.
Corresponding Author: Kadir Turan, Department of Pharmaceutical Biotechnology, Faculty of Pharmacy, University of Marmara, Haydarpasa, Kadikoy, Istanbul 34668, Turkey. kadirturan@marmara.edu.tr
Published by the Canadian Society for Pharmaceutical Sciences.
Copyright © 1998 by the Canadian Society for Pharmaceutical Sciences.
http://www.ualberta.ca/~csps