J Pharm Pharmaceut Sci (www.cspscanada.org) 9(1):124-132, 2006
Optimization of a two-step desolvation method for preparing gelatin nanoparticles and cell uptake studies in 143B osteosarcoma cancer cells
Shirzad Azarmia,b,c, Yuan Huanga, Hua Chend, Steve McQuarriea, Douglas Abramse, Wilson Road, Warren H. Finlayf, Gerald G. Millera, Raimar Löbenberga
a
Faculty of Pharmacy and
Pharmaceutical Sciences,
b
Faculty of Pharmacy,
c
d
Department of Radiation
Oncology, Cross Cancer Institute,
e
Edmonton Radiopharmaceutical Center, Cross Cancer Institute,
f
Department of Mechanical Engineering,
Received 1 November 2005, Revised 24 February 2006, Accepted 27 February 2006, Published 16 March 2006
Corresponding
Author:
Raimar Löbenberg, Faculty of Pharmacy and
Pharmaceutical Sciences,
ABSTRACT PURPOSE: To establish a matrix of parameters to synthesize nanoparticles of different sizes and to investigate the cellular uptake of these nanoparticles by osteosarcoma cancer cells in order to investigate their potential as therapeutic drug-delivery carriers. METHODS: Gelatin A and B were used to synthesize nanoparticles by a two-step desolvation process. Different parameters were investigated, including temperature, pH, concentration of glutaraldehyde, type of desolvating agent and nature of gelatin. For cell uptake studies, Texas Red labeled nanoparticles were incubated with 143B osteosarcoma cells and then evaluated using confocal laser scanning microscopy (CLSM). RESULTS: The systematic investigation of the synthesis parameters showed that it is possible to prepare gelatin-based nanoparticles with different particle sizes and a narrow size distribution. Temperature and nature of the gelatin were the most important synthesis factors. Bioimaging using CLSM showed uptake of the nanoparticles by 143B osteosarcoma cancer cells. CONCLUSIONS: Osteosarcoma cancer cells take up gelatin nanoparticles. This might improve the clinical effectiveness of anti-cancer treatments if nanoparticles are used as a drug delivery system and has important implications for future cancer treatment strategies.
INTRODUCTION
Drug targeting technology represents one of the frontier areas of science, which involves a multidisciplinary scientific approach with great potential to positively impact our health care. Drug targeting of a therapeutic moiety to the site of drug action within the body offers numerous advantages compared to the use of conventional dosage forms, including improved efficacy, reduced toxicity and overcoming drug resistance (1, 2).
Different nano-sized carriers, such as nanoparticles (3-7), polymeric micelles (8), liposomes (9), surface-modified nanoparticles (10, 11) and solid lipid nanoparticles (12), have been developed and suggested for achieving these goals.
Biodegradable nanoparticles can be synthesized from selected natural or synthetic macromolecules, such as serum albumin, polycyanoacrylates, polylactic-co-glycolic acid, and recently chitosan. Several researchers have investigated the use of gelatin as biomaterial to synthesize drug delivery systems. Gelatin nanoparticles have been used for delivery of different drugs (13-15), gene delivery (16-18), as carriers to deliver drug to lungs (19), and recently antibody modified gelatin nanoparticles were used to target lymphocytes (20), leukemic cells and primary T-lymphocytes (21).
It has been shown that particle size is an important factor for the tissue and organ distribution of nanoparticles. For example, body distribution studies have shown that nanoparticles larger than 230 nm accumulate in the spleen due to the capillary size in this organ (22). Different in vitro studies indicate that the particle size also influences the cellular uptake of nanoparticles (23-25).
The formation of gelatin-based nanoparticles has not been extensively investigated even though its first use as a base for nanoparticles was described more than 25 years ago (26). One reason for this might be that the use of native gelatin generally produces large particles with a wide size range. However, using a two-step desolvation process has been shown to be more efficient in the formation of smaller nanoparticles (27).
Gelatin is a naturally occurring polymer with low antigenicity (28, 29) and is used clinically as a plasma expander (30). Gelatin is obtained by controlled hydrolysis of the fibrous, insoluble protein, collagen, which is widely found as the major component of skin, bones and connective tissue (29). Two different gelatins, A and B with different isoelectric points (IEP), are formed following either acid or base hydrolysis, respectively. The different IEP of gelatin A and B can be used to form stable microparticles using a pH induced coacervation process (31).
Characteristic
features of gelatin are the high content of the amino acids glycine, proline
(mainly as hydroxyproline) and alanine. Gelatin molecules contain repeating
sequences of glycine, proline and alanine amino acid triplets, which are
responsible for the triple helical structure of gelatin. Commercial gelatins
are heterogeneous protein mixtures of polypeptide chains and have a wide range
of molecular weight ranging from a few thousand up to several hundred thousand
In this work we studied the effect of different preparative techniques used to synthesize gelatin-based nanoparticles. Our goal was to establish a matrix of parameters to synthesize nanoparticles of different sizes and then to investigate the cellular uptake of these nanoparticles by osteosarcoma cancer cells in order to investigate their potential in a therapeutic drug-delivery role.
MATERIALS
AND METHODS
Materials
Gelatin type A from porcine skin (175 Bloom), gelatin type B
from bovine skin (225 Bloom), glutaraldehyde grade I 25% aqueous solution,
sulforhodamine 101 acid chloride (Texas Red), and 2,4,6-trinitrobenzenesulfonic
acid (TNBS) were obtained from Sigma Chemical Co (St Louis, MO, USA). Acetone,
ethyl alcohol and acetonitrile were purchased from Caleda (
Preparation of nanoparticles
Gelatin nanoparticles were prepared by a two-step desolvation
method, previously described by Coester et al (27). In brief: 1.25 g gelatin
was dissolved in 25 mL distilled water under constant heating temperature
range. 25 mL acetone or ethanol was added to the gelatin solution as a
desolvating agent to precipitate the high molecular weight (HMW) gelatin. The
supernatant was discarded and the HMW gelatin re-dissolved by adding 25 mL
distilled water and stirring at 600 rpm under constant heating. The pH of the
gelatin solution was adjusted to values between 2.5 and 12. Acetone or ethanol
(75 mL) were added drop-wise to form nanoparticles. At the end of the process,
250 μL of 25% glutaraldehyde solution was used for preparing nanoparticles
as a cross-linking agent, and stirred for 12h at 600 rpm. Different amounts of
glutaraldehyde solution (100-500 mL) were used to determine the impact of the cross-linker
on the particle size. The remaining organic solvent was evaporated using a
rotary evaporator (IKA,
Fluorescence labeling of
nanoparticles
After the second desolvation step (see 2.2), 250 mL of a 1 mg/mL solution of Texas Red (sulforhodamine 101 acid chloride) in acetonitrile was added to the nanoparticle suspension and stirred at 600 rpm for 1 hr at 40°C. Then, the particles were cross-linked and purified as previously described.
Purification of nanoparticles
The particles were purified from synthesis residue and
unbound dyes using a size exclusion method. 1 mL of nanoparticle suspension was
injected onto a 20 mm x 35 cm Sephadex G50 column (Pharmacia LKB,
Characterization of nanoparticles
The particle size of the nanoparticles was determined by
photon correlation spectroscopy using a Zetasizer, model HSA 3000 (Malvern,
The surface charge of the nanoparticles was determined by laser Doppler anemometry using a Zetasizer, model HSA 3000. For the measurement, 100 μL of nanoparticle suspension was diluted to 4 mL with 10 mM NaCl solution followed by adjusting the pH to values between 5.0 and 8.0, using 0.25 N HCl or 0.25 N NaOH. An electric field of 150 mV was applied to observe the electrophoretic velocity of the particles. The NaCl solution compensated for the conductivity effect resulting from the addition of HCl or NaOH. All the measurements were made at room temperature, in triplicate.
Quantification of free amino
groups after cross-linking
Free amino groups were
determined using the reaction of 2, 4, 6-trinitrobenzenesulfonic acid (TNBS)
with amino groups of the particles as described by Fields (34). The
nanoparticles were washed and centrifuged four times (Airfuge, Beckman,
Cell culture
143B osteosarcoma cancer cell line (ATCC
CRL-8303) was used for the
cell uptake studies. These cells are epithelial-like adherent cells, which grow
as a monolayer. ATCC complete growth medium, minimum essential medium (Eagle)
in Earle's BSS with 0.015 mg/ml 5-bromo-2'-deoxyuridine, 90%; fetal bovine
serum, 10%, was used as the growth medium. The cells were grown in 75mL flasks
(
143B cell uptake
Approximately 104 cells were grown in a Lab-Tek
II Chamber SlideTM System (
Statistical analysis
A paired t-test was used to compare the different nanoparticle batches. The zeta potential graphs were compared using the Chi-square test with significance deemed at p=0.05.
Confocal laser scanning microscopy (CLSM)
For imaging the cells, a Zeiss LSM 510 confocal microscope (
Measurement of the intensity of fluorescence in CLSM images
The cell uptake of nanoparticles (gelatin B) with different
particle sizes (190, 283 and 330 nm) was determined and the intensity of
fluorescence in CLSM images was measured using Metamorph software version 6.2.6
(Universal Imaging Corp.,
RESULTS
AND DISCUSSION
A method of preparing gelatin nanoparticles by two-step desolvation method has been described by Coester et al (27). In our experiments, we studied the effects of varying production parameters on the nanoparticle properties. Different synthesis parameters were changed, including temperature, pH, concentration of glutaraldehyde, type of desolvating agent and nature of gelatin. The goal was to prepare small nanoparticles with a narrow size distribution. It has been shown that particle size has a great impact on the uptake of nanoparticles. Desai and co-workers (23) showed that 100 nm size nanoparticles had 2.5 fold greater uptake compared to 1 µm and 6 fold higher uptake compared to 10 µm microparticles in a Caco-2 cell line.
The results of other researchers also showed that particle size significantly affects cellular and tissue uptake, and, in some cell lines, only the submicron size particles are taken up efficiently in lieu of the larger size microparticles (24, 25).
We investigated the effect of these different parameters on the particle size and the polydispersity index, where the polydispersity index measures the second moment of the size distribution of the nanoparticle population. A lower polydispersity index indicates a narrower size distribution.
To study the effect of temperature on the particle size of the nanoparticles, only the temperature was changed in the experiments and all other parameters were kept constant. Acetone was used as desolvating agent (75 mL) and glutaraldehyde (250 µL) as cross-linker. The results are shown in Table 1.
Table 1. Influence of the temperature and gelatin type on nanoparticle size
(n=3). (Acetone was used as desolvating agent and 250 µL of
glutaraldehyde as cross-linking agent)
|
Size (nm) |
Polydispersity
Index |
Type of
gelatin |
Temperature
(°C) |
|
163±24 |
0.061±0.007 |
A |
40 |
|
112±21 |
0.0915±0.031 |
B |
40 |
|
252±53 |
0.0202±0.012 |
A |
50 |
|
214±31 |
0.026±0.015 |
B |
50 |
|
306±54 |
0.0674±0.042 |
A |
60 |
|
303±52 |
0.0695±0.048 |
B |
60 |
During these investigations, it was found that the preparation of nanoparticles at ambient temperature (25 °C) was not possible because the gelatin formed a highly viscous gel at this temperature. The results at 40, 50 and 60°C showed that temperature has an impact on the particles size. The smallest nanoparticles were prepared at 40°C with gelatin A or B. Increasing the temperature to 50 and 60 °C increased the particle size. This might be explained by the gelling properties of gelatin. In solution, the triple helical structure begins to uncoil when the temperature increases. The viscosity simultaneously decreases. At 40°C, the chains seem to be sufficiently uncoiled and the addition of the desolvating agent causes a better controlled precipitation of the macromolecules compared to higher temperatures.
The nature of gelatin also had an effect on the particle characteristics. At 40 and 50°C nanoparticles made from gelatin A were generally larger compared to nanoparticles made from gelatin B (Table 1) but were of similar size when prepared at 60°C. In addition the polydispersity of the particle size distribution was narrower for gelatin A compared to gelatin B.
The isoelectric points of gelatin A and B are approximately 6.1 and 4.5, respectively. To form nanoparticles in the second desolvation step, the pH of the gelatin solution has to be adjusted away from their isoelectric points to either pH 2.5 or 12. The addition of the desolvating agent reduces the water available to keep the gelatin in solution resulting in shrinkage of the hydrated gelatin chains. At a certain point the hydration is too low and the protein chains precipitate as nanoparticles. The results using gelatin A and B showed that pH 2.5 was the optimum pH for preparing the nanoparticles. Increasing the pH to 4 or higher caused early agglomeration of the gelatin when the desolvating agent was added. A possible explanation of this observation is that at pH 2.5 or 12 protein chains are highly positively or negatively charged. The electrostatic repulsion prevents the polymer chains from uncontrolled agglomeration. After the nanoparticles are formed, their surface has a sufficient zeta potential to prevent further agglomeration of the particles.
To study the effect of the concentration of glutaraldehyde as a cross-linking agent, 200, 300, 400 and 500 mL aliquots of a 25% v/v aqueous glutaraldehyde solution were added to the nanoparticles. The nanoparticles were prepared at 50°C using gelatin B under the conditions described above. In these experiments, acetone was used as the desolvating agent.
These amounts of the cross-linker were sufficient to stabilize the particles. Lower amounts were not sufficient to cross link the nanoparticles because a steep increase in the particle size was observed upon storage (data not shown). This can be attributed to the swelling of the gelatin in aqueous media after the organic solvent was removed. Increasing the concentration of glutaraldehyde from 200 to 500 mL did not show any significant effect on the particle size of the nanoparticles (p>0.05), as shown in Table 2.
Table 2.
Influence of the cross-linker concentration on the particle size (n=3). (Acetone was used as desolvating agent for gelatin B at 50°C temperature)
|
Glutaraldehyde volume (mL) |
Size (nm) |
Polydispersity
Index |
|
200 |
197±15 |
0.0477±0.037 |
|
300 |
219±20 |
0.0785±0.042 |
|
400 |
182±11 |
0.0121±0.019 |
|
500 |
197±8 |
0.0671±0.024 |
The determination of free amino groups on the surface of nanoparticles cross-linked using 250 mL of glutaraldehyde showed that about 12% of the amino groups were still available. This is in accordance with results reported by Weber et al. and Rubino et al (33, 35). The narrow size distribution of the nanoparticles indicates that the glutaraldehyde reacted with amino groups on the same particle rather than linking two particles together. This might be due to the zeta potential of the particles, which prevents them from coming close together. The degree of cross-linking and therefore the degree of free amino groups are important factors because they have an impact on the biodegradability of the particles as described by various groups (36-38). Additionally, free amino groups can be used to link drugs or other active principals like antibodies to the nanoparticle surface. Further studies on the degradation kinetics of the gelatin nanoparticles are necessary in order to estimate their intracellular half-life.
To evaluate the effects of desolvating agent type, acetone and ethyl alcohol were employed. In these experiments the temperature was kept constant at 50 °C and 250 µL of glutaraldehyde was added as cross-linking agent. The results are shown in Table 3.
The results show that acetone was the preferred desolvating agent as the nanoparticles prepared with acetone were generally smaller and had a lower polydispersity index compared to those prepared with ethyl alcohol. The zeta potential of the gelatin A and B nanoparticles was measured in a pH range of 2 to 12 using a zeta sizer as shown in Figure 1.
Table 3.
Influence of type of desolvating agent on the particle size (n=3). (250 µL of glutaraldehyde was used as
cross-linking agent at 50°C temperature)
|
Gelatin |
Desolvating
agent |
Size (nm) |
|
A |
Acetone |
228 ±11 |
|
A |
Ethanol |
386 ±8 |
|
B |
Acetone |
185 ±13 |
|
B |
Ethanol |
286 ±10 |
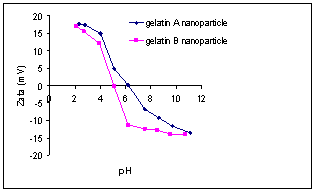
Figure
1.
The
effect of pH on the zeta potential of gelatin A and gelatin B nanoparticles.
Increasing the pH of the nanoparticle suspension led to a
decrease in the zeta potential. As shown in Figure 1, the zeta potential was a
function of the nature of the gelatin used and the pH of the medium. Positively
or negatively charged particles can be obtained between pH 4 and 6. However,
the zeta potential of gelatin B reached a plateau, after passing pH 6.5 where
the change in the zeta potential became less pronounced relative to the change
between pH 4 to pH 6. This plateau was not as pronounced with gelatin A nanoparticles. Labeling the nanoparticles with
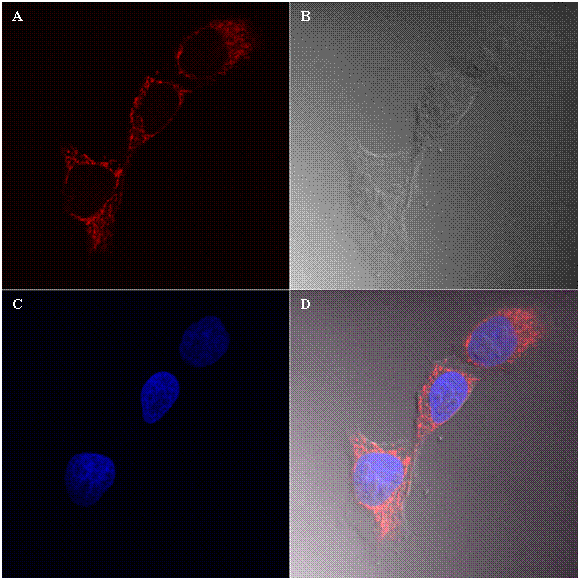
Figure 2 shows the cellular uptake and distribution of nanoparticles with an average particle size of 190 nm by 143B cells. In this figure, A shows the detection of Texas red (Red channel), B shows bright field, part C is the detection of DAPI a nucleus staining dye and part D is the overlay of all three images. As shown in Figure 2 A, the labeled nanoparticles were found mainly in the cytoplasm of cancer cells.