Tom Hobman

Tom Hobman
Ph.D., University of British Columbia
Professor
Office: 6-142 Katz BuildingLaboratory: 6-120 Katz Building
Telephone: 780-492-6485
tom.hobman@ualberta.ca
Awards
- Canada Research Chair in RNA Viruses and Host Interactions (2010-2018)
- Fellow Canadian Academy of Health Sciences (2021)
Research Interests
There are two major projects in the laboratory
Project I: RNA virus host interactions: RNA virus infections impose huge economic and social burdens around the globe. Direct-acting antiviral therapeutics and vaccines can be highly effective in controlling epidemic and pandemic viruses, but these drugs/prophylactics take significant time to develop and their efficacy is often limited to specific viral strains. To minimize the impact of future emerging RNA viruses, a stockpile of broad-spectrum antivirals is required to serve as a first line of defense against these pathogens. Identification of host cell-based pathways that are exploited or inhibited by multiple viruses is expected to reveal novel targets for antiviral therapy. Using targeted and unbiased screening approaches, we have identified multiple host cell pathways that can be pharmacologically inhibited or enhanced to reduce infection by multiple RNA viruses including SARS-CoV-2, the causative agent of COVID-19. In addition to SARS-CoV-2 and other coronaviruses, our lab studies HIV and pathogenic mosquito-transmitted viruses such as West Nile virus, Dengue virus, Zika virus, Mayaro virus and Chikungunya virus.
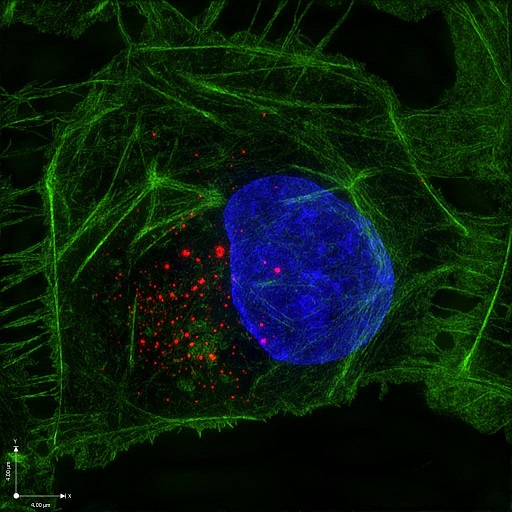 |
| Super-resolution image of Zika virus replication complex (red) in mammalian cell 24 hours post-infection |
Project II: Regulation of RNA interference pathways and breast cancer: Regulation by miRNAs and the RNAi pathway affects expression of more than 60-70% of mammalian genes. Accordingly, deregulation of this pathway could alter the expression of a vast array of cellular gene products, ultimately changing the phenotypic properties of that cell (i.e. metastatic transformation). Members of the Argonaute family of proteins bind the miRNA to form silencing effector complexes. Ago2 has endonuclease activity and is the most abundant and ubiquitously expressed Argonaute isoform. Our research is focused on understanding how Ago2 activity is regulated under normal and disease conditions. Emerging data suggest that Ago2 is an important tumor suppressor in breast cancer.
Our research is supported by: Canadian Institutes of Health Research, Natural Sciences and Engineering Research Council and Tonix Pharmaceuticals.
Selected Publications
Name of trainees are underlined
Airo, A.M., Felix-Lopez, A., Mancinelli, V., Evseev, D., Paskowski, P., Lopez-Orozco, J., Shire, K., Frappier, L., Magor, K.E. and Hobman, T.C. (2022) Flavirus capsids inhibit the interferon response. Viruses 14(5):968. doi: 10.3390/v14050968
Doan, M., Roczkowsky, A., Smith, M, Blevins, G., van Landeghem, F., Gelman, B., Branton, W., Stapleton, J., Hobman, T. and Power, C. (2021) Infection of glia by human pegivirus suppresses peroxisomal and antiviral signaling pathways. J Virol 95(23):e0107421. doi: 10.1128/JVI.01074-21. Epub 2021 Sep 15.
Nguyen, L., McCord, K.A., Bui, D.T., Bouwman, K.M., Kitova, E.N., Elaish, M., Kumawat, D., Daskhan, D.C., Tomris, I., Han, L., Chopra, P., Yang, T-J., Willows, S.D., Mason, A.L., Mahal, L., Lowary, T.L., West, L.J., Hsu, S-T.D., Hobman, T.C., Tompkins, S.M., Boons, G-J., de Vries, R.P., Macauley, M.S. and Klassen, J.S. (2021) Sialic acid-Dependent Binding and Viral Entry of SARS-CoV-2. Nat Chem Biol 18(1) 81-90. doi: 10.1038/s41589-021-00924-1.
Kumar, A., Ishida, R., Strilets, T., Cole, J., Lopez-Orozco, J., Fayad, N., Felix-Lopez, A., Elaish, M., Evseev, D., Magor, K.E., Mahal, L.K., Nagata, L.P., Evans, D.H. and Hobman, T.C. (2021) SARS-CoV-2 non-structural protein 1 inhibits the induction and signalling of type I interferons by depleting key signalling factors. J. Virol June 10; 95(13): e0026621. doi: 10.1128/JVI.00266-21
Knobloch, B., Ishida, R., Hobman, T.C. and Rachubinski, R.A. (2021) Peroxisomes exhibit compromised structure and matrix protein content in SARS-CoV-2-infected cells. Mol Biol Cell, July 1;32(14):1273-1282. doi: 10.1091/mbc.E21-02-0074. Epub 2021 May 19.
Limonta, D., Dagman, L.D., Branton, W., Makio, T., Wozniak, R.W., Power, C. and Hobman, T.C. (2021) Nodosome inhibition as a novel broad-spectrum antiviral strategy against arboviruses and SARS-CoV-2. Antimicrob Agents Chem May 17:AAC.00491-21. doi: 10.1128/AAC.00491-21
Zhang, K., Miorin, L., Makio, T., Dehghan, I., Gao, S., Xie, Y., Zhong, H., Esparza, M., Kehrer, T., Kumar, A., Hobman, T.C., Ptak, C., Gao, B., Minna, J.D., Chen, Z., Garcia-Sastre, A., Ren, Y., Wozniak, R.W. and Fontoura, B.M.A. (2021) Nsp1 Protein of SARS-CoV-2 Disrupts the mRNA Export Machinery to Inhibit Host Gene Expression. Science Advances Feb 5;7(6):eabe7386. doi: 10.1126/sciadv.abe7386
Bernard, I., Limonta, D., Mahal, L.K. and Hobman, T.C. (2020) Endothelium Infection and Dysregulation by SARS-CoV-2: Evidence and Caveats in COVID-19. Viruses 2020 Dec 26;13(1):E29. doi: 10.3390/v13010029.
Glasgow, A., Glasgow, J., Limonta, D., Solomon, P., Lui, I., Zhang, Y., Nix, M.A., Rettko, N.J., Zha, S., Yamin, R., Kao, K., Rosenberg, O.S., Ravetch, J.V., Wiita, A.P., Leung, K.K., Lim, S.A., Zhou, X.X., Hobman, T.C., Kortemme, T. and Wells, J.A. (2020) Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc Natl Acad Sci Nov 10;117(45):28046-28055. doi: 10.1073/pnas.2016093117. Epub 2020 Oct 22
Griffiths, C.D., Bilawchuk, L.M., McDonough, J.E., Jamieson, J.C., Elawar, F., Cen, Y., Duan, W., Lin, C., Song, H., Casanova, J-L., Ogg, S., Jensen, L.D., Thienpont, B., Victoria-Ansalem, A., Hobman, T.C., Proud, D., Moraes, T.J., and Marchant, D.J. (2020) IGF1R is a Respiratory Syncytial Virus Entry Receptor: Viral recruitment of its entry coreceptor from the nucleus to the cell surface Nature Jul;583(7817):615-619. doi: 10.1038/s41586-020-2369-7. Epub 2020 Jun 3
Xu, Z., Lodge, R., Power, C., Cohen, E.A. and Hobman, T.C. (2020) The HIV-1 accessory protein Vpu downregulates peroxisome biogenesis. mBio Mar 3;11(2). pii: e03395-19. doi: 10.1128/mBio.03395-19.
Limonta, D., Jovel, J., Kumar, A., Liu, J., Hou, S., Airo, A.M., Lopez-Orozco, J., Wong, C.P., Saito, L., Branton, W., Wong, G. K-S., Mason, A., Power, C. and Hobman, T.C. (2019) Fibroblast growth factor 2 enhances Zika virus infection and facilitates persistence in human fetal brain. J Infect Dis Feb 13. 220(8):1377-1387. pii: jiz073. doi: 10.1093/infdis/jiz073.
Wong, C.P., Xu, Z, Hou, S., Limonta, D., Kumar, A., Power, C. and Hobman, T.C. (2019) Interplay between Zika virus and peroxisomes during infection. Cells Jul 15;8(7). pii: E725. doi: 10.3390/cells8070725
Kumar, A., Jovel, J., Lopez-Orozco, J., Limonta, D., Airo, A.M., Hou, S., Stryapunina, I., Fibke, C., Moore, R.B. and Hobman, T.C. (2018) Human Sertoli cells support high levels of Zika virus replication and persistence Scientific Reports. 8: 5477. doi: 10.1038/s41598-018-23899-x.
Hou, S., Kumar, A., Airo, A.M., Xu, Z., Stryapunina, I., Wong, C.P., Branton, W., Tchesnokov, E., Gotte, M., Power, C., Hobman, T.C. (2017) Zika virus employs multiple strategies to hijack stress and modulate host stress response. J Virol91: 1-29.
This article was selected by the editors for inclusion in "Spotlight," a feature in the Journal that highlights research articles of significant interest from the current issue.
Xu, Z., Asahchop, E., Branton, W.G., Gelman, B.B., Power, C. and Hobman, T.C. (2017) MicroRNAs upregulated during HIV infection target peroxisome biogenesis factors: Implications for virus biology, disease mechanisms and neuropathology. PLoS Pathogens Jun 8;13(6):e1006360. doi: 10.1371/journal.ppat.1006360.
Reid, C.R. and Hobman, T.C. (2017) The nucleolar helicase DDX56 redistributes to West Nile virus assembly sites. Virology 500:169-177. doi: 10.1016/j.virol.2016.10.025.
Kumar, A., Hou, S., Airo, A.M., Limonta, D., Mancinelli, V., Branton, W., Power, C., Hobman, T.C. (2016) Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep 17 (12):1766-1775.
You, J., Hou, S., Malik-Soni, N., Xu, Z., Kumar, A., Rachubinski, R.A., Frappier, L., and Hobman, T.C. (2015) Flavivirus infection impairs peroxisome biogenesis and early anti-viral signaling.J. Virol 89: 12349-61. doi: 10.1128/JVI.01365-15.
Selected by editors for inclusion in "Spotlight," a feature in the Journal that highlights research articles of significant interest from the current issue.
Singaravelu, R, O'Hara, Shifawn, Jones, DM. Chen, R., Steenbergen, R.H., Kumar, A, Taylor, N.G., Srinivasan, P., Lyn, R.K., Quan, C., Özcelik, D., Nguyen, M-A., Rayner, K.J., Hobman, T., Tyrrell, D.L., Russell, R.S., and Pezacki, J.P. (2015) Oxysterol induced microRNAs regulate antiviral responseNature Chem Biol11: 988-93. doi: 10.1038/nchembio.1940.
Lopez-Orozco, J., Pare, J.M., Holme, A.L., Chaulk, S.G., Fahlman, R.P. and Hobman, T.C. (2015) Functional analyses of phosphorylation events in human Argonaute 2.RNA 21: 2030-8. doi: 10.1261/rna.053207.115.
Willows, S., Ilkow, C.S. and Hobman, T.C. (2014) Phosphorylation and membrane-association of the Rubella virus capsid protein is important for its anti-apoptotic function. Cell Micro16: 1201-10.
Mangala Prasad, V., Willows, S.D., Fokine, A., Battisti, A. J., Sun, S., Plevka, P., Hobman, T.C.,Rossmann, M.G. (2013) Rubella virus capsid protein structure and its role in virus assembly and infection. Proc. Natl. Acad. Sci.110: 20105-20110.
Wang, Y., Mercier, R., Hobman, T.C., and LaPointe, P. (2013) Regulation of RNA interference by Hsp90 is an evolutionarily conserved process. Biochim Biophys Acta - Mol Cell Res 1833: 2673-2681.
Urbanowski, M.D. and Hobman, T.C. (2013) The West Nile Virus Capsid Protein Blocks Apoptosis through a Phosphatidylinositol 3-kinase-dependent Mechanism.J. Virol.87: 872-881.
Pare, J.M., LaPointe, P. and Hobman, T.C. (2013) Hsp90 co-chaperones p23 and FKBP4 physically interact with hAgo2 and activate RNAi-mediated silencing in mammalian cells. Mol Biol Cell24: 2303-2310.
Laboratory Members
Graduate Students
Alberto Felix-Lopez
Postdoctoral Fellow
Dr. Mohamed Elaish
Research Associates
Dr. Joaquín López-Orozco
Dr. Zaikun Xu
Undergraduate Research Students
Karen He
Technicians
Nawell Fayad
Valeria Mancinelli
Eileen Reklow